Note
Click here to download the full example code
2D profiles stored in core_profiles ids¶
Out:
loading equilibrium
loading core_profile
WARNING: density profile for neutral species Be is equal to 0 in core_profiles IDS skipping this species.
WARNING: density profile for neutral species Ne is equal to 0 in core_profiles IDS skipping this species.
Sampling psi...
Plotting...
import numpy as np
from matplotlib import pyplot as plt
from cherab.core.math import sample2d, sample3d
from cherab.core.atomic import deuterium
from cherab.imas import EquilibriumIDS, CoreProfilesIDS
# IDS Setting
SHOT = 134000
RUN = 27
USER = "koyo"
DATABASE = "iter"
print("loading equilibrium")
equilibrium_ids = EquilibriumIDS(SHOT, RUN, user=USER, database=DATABASE)
imas_equilibrium = equilibrium_ids.time(-1)
print("loading core_profile")
core_profile_ids = CoreProfilesIDS(SHOT, RUN, user=USER, database=DATABASE)
core_plasma = core_profile_ids.create_plasma(imas_equilibrium)
# plot equilibrium
rmin, rmax = imas_equilibrium.r_range
zmin, zmax = imas_equilibrium.z_range
# sample every 5 cm
resolution = 0.05
nr = int(round((rmax - rmin) / resolution))
nz = int(round((zmax - zmin) / resolution))
print("Sampling psi...")
r, z, psi_sampled = sample2d(imas_equilibrium.psi_normalised, (rmin, rmax, nr), (zmin, zmax, nz))
print("Plotting...")
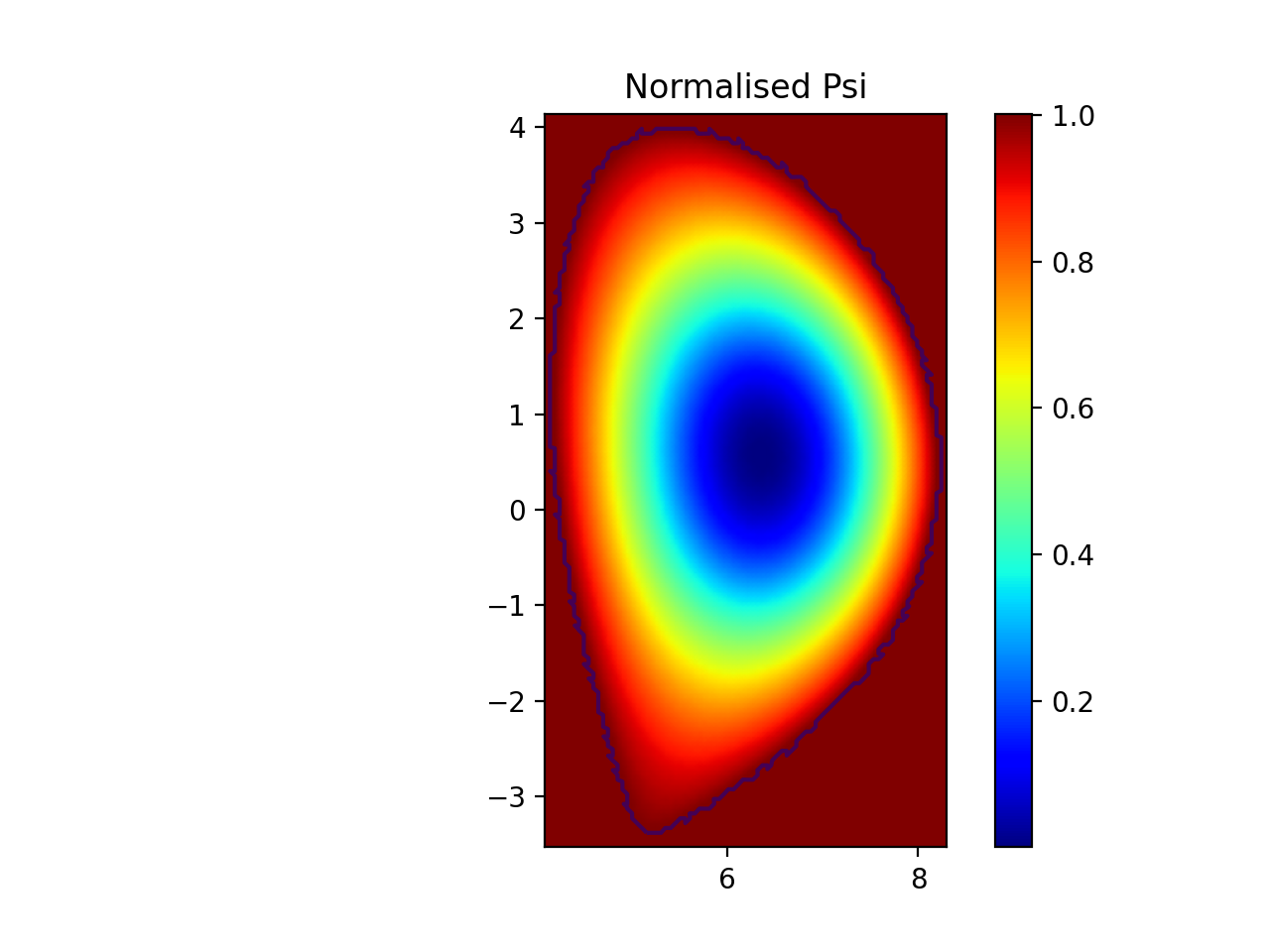
plt.figure()
plt.axes(aspect="equal")
plt.pcolormesh(r, z, np.transpose(psi_sampled), cmap="jet", shading="gouraud")
plt.autoscale(tight=True)
plt.colorbar()
plt.contour(r, z, np.transpose(psi_sampled), levels=[1.0])
plt.title("Normalised Psi")
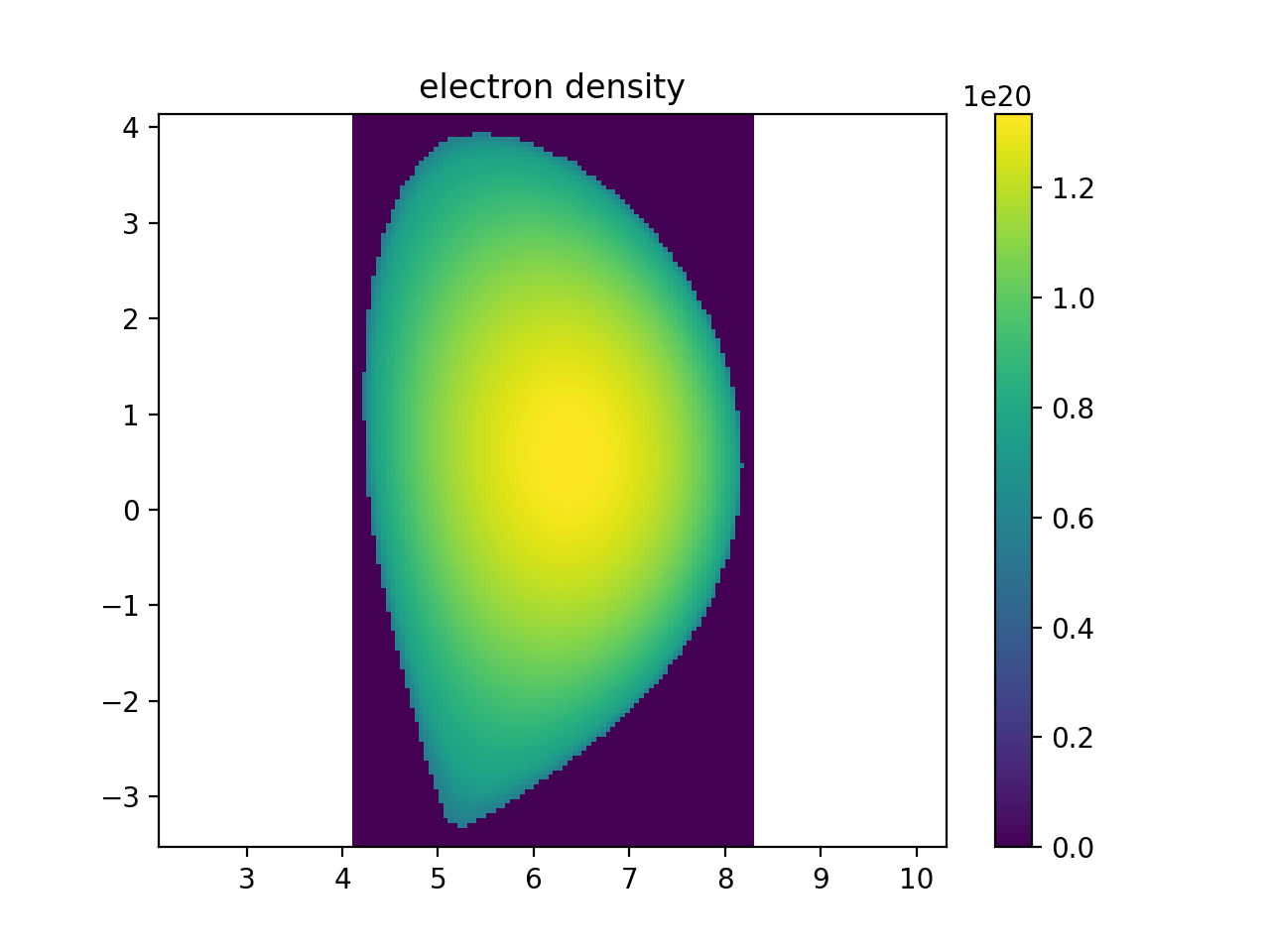
plt.figure()
r, _, z, ne_samples = sample3d(core_plasma.electron_distribution.density, (rmin, rmax, nr), (0, 0, 1), (zmin, zmax, nz))
plt.imshow(np.transpose(np.squeeze(ne_samples)), extent=[rmin, rmax, zmin, zmax], origin="lower")
plt.colorbar()
plt.axis("equal")
plt.xlim(rmin, rmax)
plt.ylim(zmin, zmax)
plt.title("electron density")
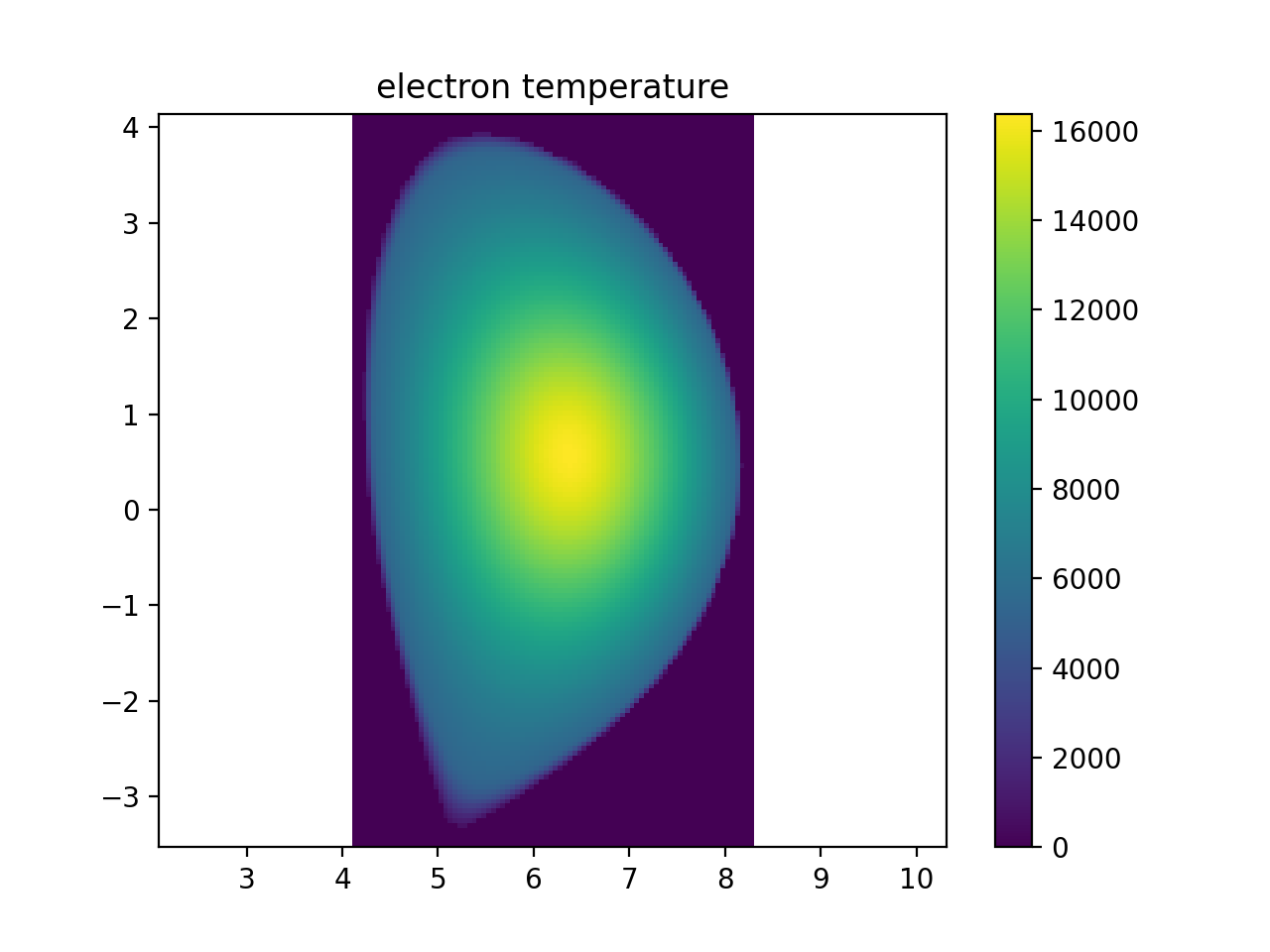
plt.figure()
r, _, z, te_samples = sample3d(
core_plasma.electron_distribution.effective_temperature, (rmin, rmax, nr), (0, 0, 1), (zmin, zmax, nz)
)
plt.imshow(np.transpose(np.squeeze(te_samples)), extent=[rmin, rmax, zmin, zmax], origin="lower")
plt.colorbar()
plt.axis("equal")
plt.xlim(rmin, rmax)
plt.ylim(zmin, zmax)
plt.title("electron temperature")
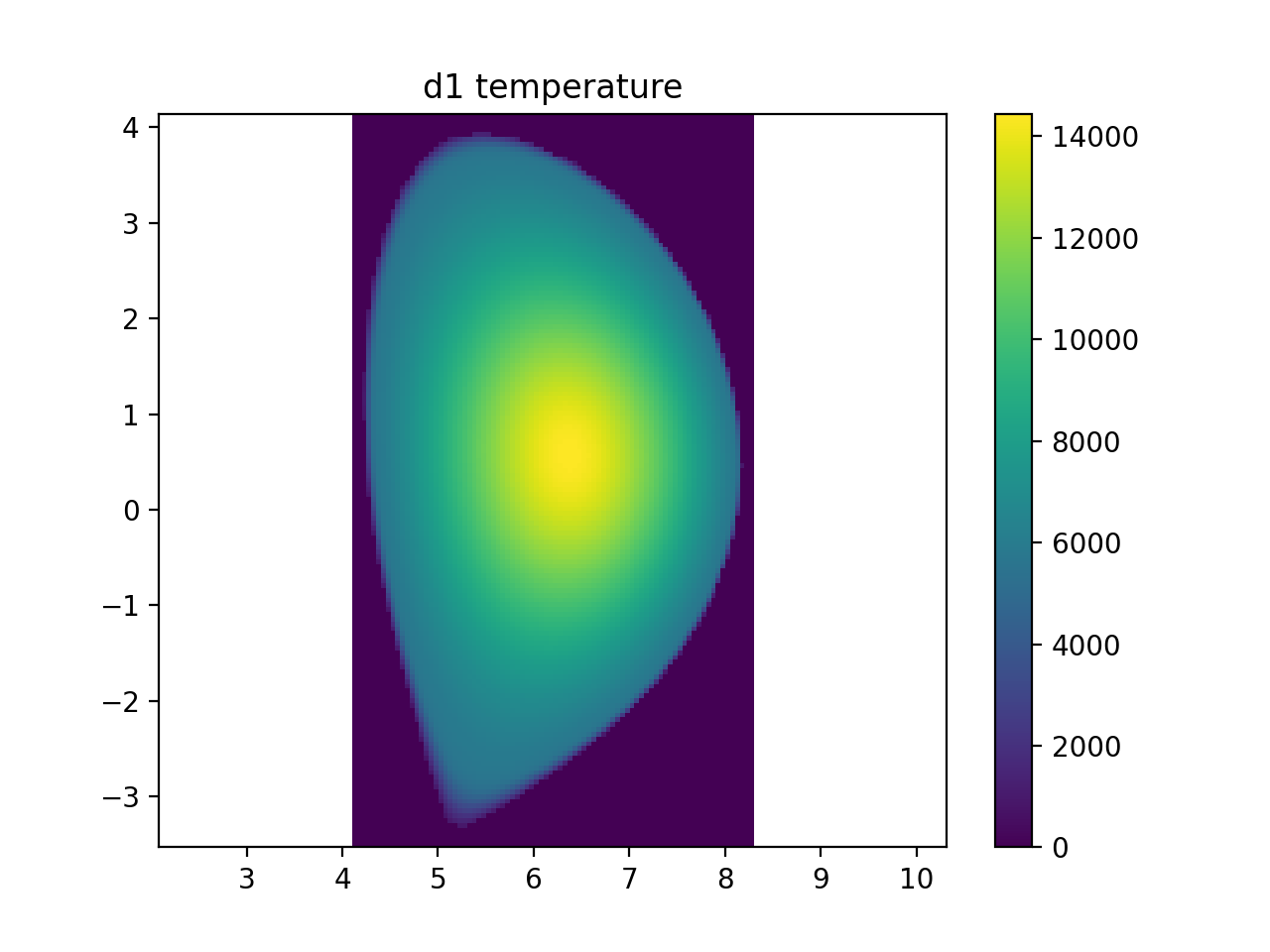
d1 = core_plasma.composition.get(deuterium, 1)
plt.figure()
r, _, z, ti_samples = sample3d(d1.distribution.effective_temperature, (rmin, rmax, nr), (0, 0, 1), (zmin, zmax, nz))
plt.imshow(np.transpose(np.squeeze(ti_samples)), extent=[rmin, rmax, zmin, zmax], origin="lower")
plt.colorbar()
plt.axis("equal")
plt.xlim(rmin, rmax)
plt.ylim(zmin, zmax)
plt.title("d1 temperature")
plt.figure()
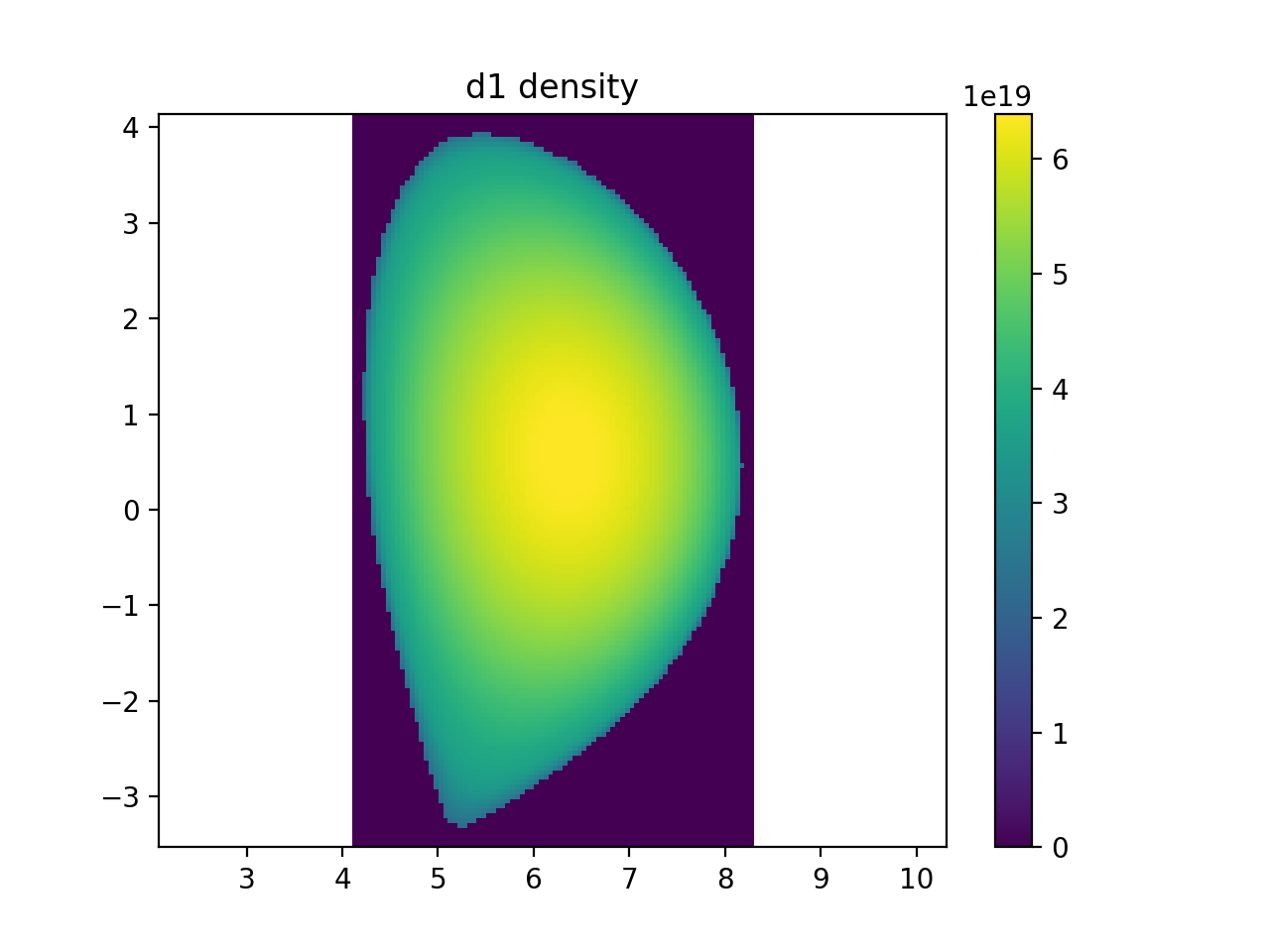
r, _, z, ni_samples = sample3d(d1.distribution.density, (rmin, rmax, nr), (0, 0, 1), (zmin, zmax, nz))
plt.imshow(np.transpose(np.squeeze(ni_samples)), extent=[rmin, rmax, zmin, zmax], origin="lower")
plt.colorbar()
plt.axis("equal")
plt.xlim(rmin, rmax)
plt.ylim(zmin, zmax)
plt.title("d1 density")
plt.figure()
d1_velocity = np.zeros((100, 100))
xrange = np.linspace(rmin, rmax, 100)
yrange = np.linspace(zmin, zmax, 100)
for i, x in enumerate(xrange):
for j, y in enumerate(yrange):
d1_velocity[j, i] = d1.distribution.bulk_velocity(x, 0.0, y).length
plt.imshow(d1_velocity, extent=[rmin, rmax, zmin, zmax])
plt.colorbar()
plt.axis("equal")
plt.xlim(rmin, rmax)
plt.ylim(zmin, zmax)
plt.title("d1 velocity")
plt.show()
Total running time of the script: ( 0 minutes 0.000 seconds)