Note
Click here to download the full example code
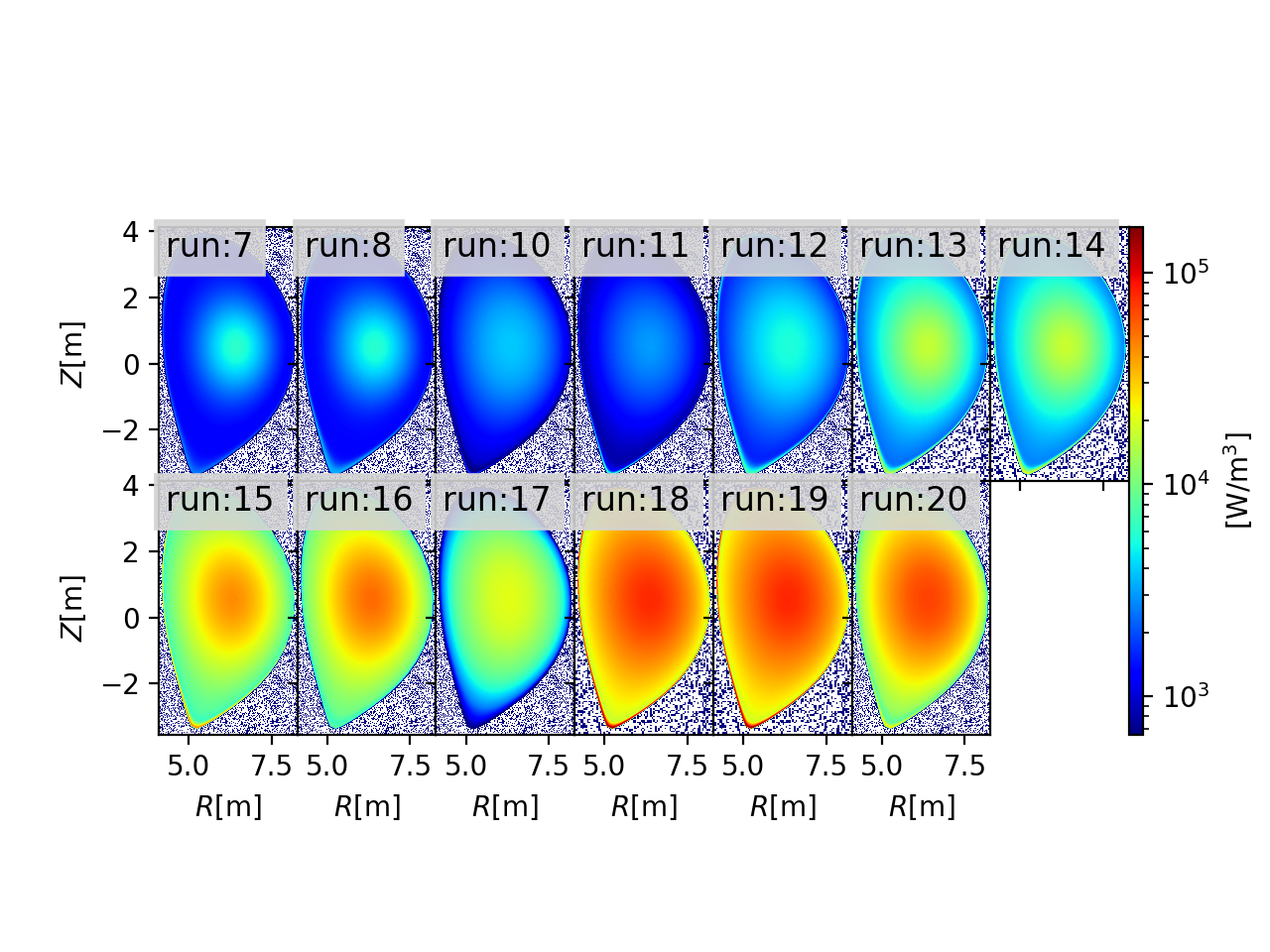
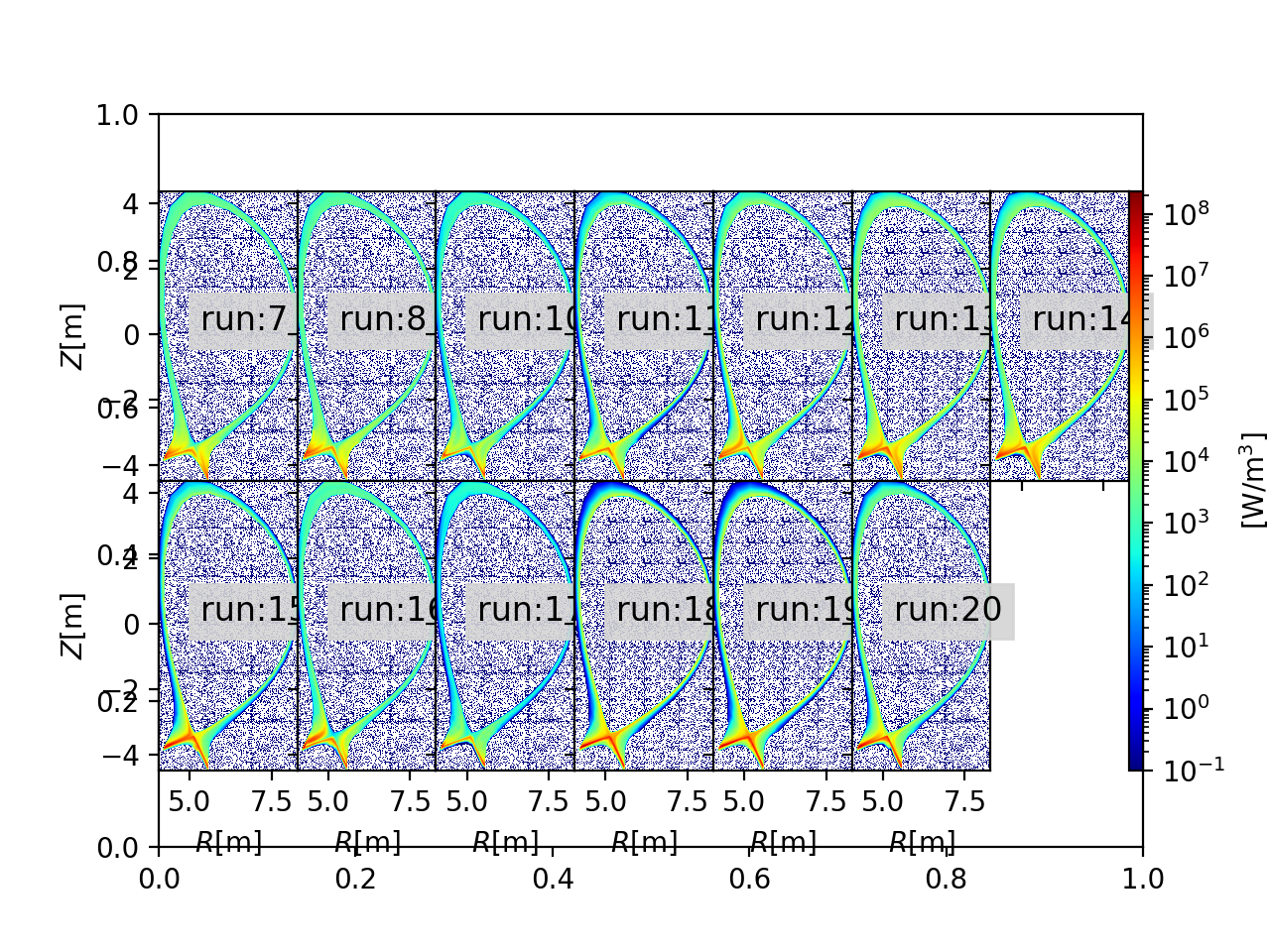
2D profiles generated by JINTRAC¶
Out:
importing shot:134000, run:7, source:'radiation', particle:'electrons'
importing shot:134000, run:8, source:'radiation', particle:'electrons'
importing shot:134000, run:10, source:'radiation', particle:'electrons'
importing shot:134000, run:11, source:'radiation', particle:'electrons'
importing shot:134000, run:12, source:'radiation', particle:'electrons'
importing shot:134000, run:13, source:'radiation', particle:'electrons'
importing shot:134000, run:14, source:'radiation', particle:'electrons'
importing shot:134000, run:15, source:'radiation', particle:'electrons'
importing shot:134000, run:16, source:'radiation', particle:'electrons'
importing shot:134000, run:17, source:'radiation', particle:'electrons'
importing shot:134000, run:18, source:'radiation', particle:'electrons'
importing shot:134000, run:19, source:'radiation', particle:'electrons'
importing shot:134000, run:20, source:'radiation', particle:'electrons'
from matplotlib import pyplot as plt
from cherab.core.math import sample2d
from cherab.imas import EquilibriumIDS, CoreSourcesIDS, EdgeSourcesIDS
from cherab.iter.tools.visualization import plot_2D_profile
# IDS Setting
SHOT = 134000
RUNs = [7, 8] + [i for i in range(10, 20 + 1)]
USER = "koyo"
DATABASE = "iter"
SOURCE = "radiation"
PARTICLE = "electrons"
data_core = []
data_edge = []
for RUN in RUNs:
print(f"importing shot:{SHOT}, run:{RUN}, source:'{SOURCE}', particle:'{PARTICLE}'")
equilibrium_ids = EquilibriumIDS(shot=SHOT, run=RUN, user=USER, database=DATABASE)
imas_equilibrium = equilibrium_ids.time(-1)
core_sources = CoreSourcesIDS(shot=SHOT, run=RUN, user=USER, database=DATABASE)
edge_sources = EdgeSourcesIDS(shot=SHOT, run=RUN, user=USER, database=DATABASE)
core_sources.energy(source=SOURCE, particle=PARTICLE, equilibrium=imas_equilibrium)
edge_sources.mesh().energy(source=SOURCE, particle=PARTICLE)
# generate 2D function
core_2D = core_sources.map2d()
edge_2D = edge_sources.map2d()
# sampling limit
core_lim = {"xlim": imas_equilibrium.r_range, "ylim": imas_equilibrium.z_range}
edge_lim = {"xlim": edge_sources.r_range, "ylim": edge_sources.z_range}
# sample every 1 cm
resolution = 0.01
nr_core = int(round((core_lim["xlim"][1] - core_lim["xlim"][0]) / resolution))
nz_core = int(round((core_lim["ylim"][1] - core_lim["ylim"][0]) / resolution))
nr_edge = int(round((edge_lim["xlim"][1] - edge_lim["xlim"][0]) / resolution))
nz_edge = int(round((edge_lim["ylim"][1] - edge_lim["ylim"][0]) / resolution))
# sampling r,z, distibution
r_core, z_core, core = sample2d(
core_2D, (*core_lim["xlim"], nr_core), (*core_lim["ylim"], nz_core)
)
r_edge, z_edge, edge = sample2d(
edge_2D, (*edge_lim["xlim"], nr_edge), (*edge_lim["ylim"], nz_edge)
)
core[core < 0] = 0
edge[edge < 0] = 0
data_core.append(core)
data_edge.append(edge)
# plot
plot_mode = "log"
ax_row = 2
fig1 = plt.figure()
fig2 = plt.figure()
plot_2D_profile(
fig=fig1,
data=data_core,
xdata=r_core,
ydata=z_core,
ax_row=ax_row,
title=[f"run:{run}" for run in RUNs],
titlekw={"fontsize": 12, "y": 0.92},
plot_mode=plot_mode,
)
plot_2D_profile(
fig=fig2,
data=data_edge,
vmin=1.0e-1,
xdata=r_edge,
ydata=z_edge,
ax_row=ax_row,
title=[f"run:{run}" for run in RUNs],
titlekw={"fontsize": 12, "x": 0.30, "y": 0.55},
plot_mode=plot_mode,
)
plt.show()
Total running time of the script: ( 0 minutes 0.000 seconds)